Bioinformatics Optimisation System for Primer-Capture-Capture-Primer Designs
| Working Group: | WG Industrial Mathematics |
| Leadership: | Prof. Dr. Dr. h.c. Peter Maaß ((0421) 218-63801, E-Mail: pmaass@math.uni-bremen.de ) |
| Processor: | Dr. Manfred Nölte |
| Project partner: | SIRS-Lab GmbH, Jena |
| Time period: | 01.07.2006 - 30.06.2008 |
Biochips are currently being used more and more in the field of medical diagnostics. With their help, the activity of genes will be identified very quickly and used for early diagnosis and therapy control.
Situation at the outset.Microarrays (also termed DNA chips ) make it possible to create and analyse biological data with a speed which has not been possible until now because the presence of thousands of genes on one microsupport facilitates a massive parallel working.
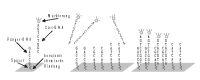
Short DNA fragments are immobilised in a structured way on a support substrate. The DNA immobilised on the matrix at a specific, i.e. known, locus is termed capture DNA. The nucleic acid to be analysed, which has a fluorescent marker, is termed the target DNA. The target DNA is identified via the locus of the hybridisation on the chip and quantified, where necessary, by the strength of the fluorescence. The loci and the intensity of the target hybridisations are analysed by means of fluorescence scanning after targets which are unspecifically bonded have been washed.
Preparatory Work. The SIRS-Lab GmbH, a biotechnology company with headquarters in Jena/Thuringia, is using DNA microarray technology to carry out research into the molecular background of acute inflammatory processes, among other things. Their objective here is to develop new biomarkers for the early diagnosis and to observe the course of infections and sepsis and to utilise them for the subsequent routine clinical diagnosis. The SIRS-Lab 1 GmbH and an independent institute founded by Bremen University have already collaborated on the design of capture libraries. At the University of Bremen, the work of the Centrum für angewandte Gensensorik (CAG) has included a dissertation "Optimisation of oligonucleotide library for DNA microarrays" and two Diplom dissertations have been completed in this field. Moreover, there is a collaboration with the ttz/BIBIS (Bremerhaven Institute for Biological Information Systems), which is presently computing capture probes as part of the "Nanoparts" EU project.
Project Objective. The optimisation of capture libraries, i.e. sets of captures, which are highly specific and highly sensitive, is a combinatorial optimisation problem. Moreover, the optimisation problem becomes even more complex if, as well as selecting suitable capture nucleic acids, primers for target preparation are selected by means of the PCR (polymerase chain reaction) method. When selecting the primers and captures, dependencies must be taken into account, for example the primers and captures, dependencies must be taken into account, for example the position on the target sequence and the separation of the primer or capture as well as the intersections of the false positive sequence correspondences. The project aims to develop a system to optimise the design of primer-capture-capture-primer experiments. For a given specification (list of the accession numbers, melting point and length of the primers and the captures, length of the DNA to be amplified, position of the capture, etc.) an optimised set of primers and captures is computed for each accession number.
Starting point for the solution.In furtherance of the above-mentioned preparatory work, a system for a parallel computer architecture based on Linux/Unix is being developed. The system components are:
- BLAST database
- tools for computing the oligonucleotide properties (primer, capture)
- combinatorial optimisation
- documentation of the computation result
The above-mentioned dependencies between the primers and the captures continue on the level of the primer-primer (P-P) and the capture-capture design (C-C design). This provides the opportunity to match or couple the C-C design specifically to one P-P design. This leads to problems which can be solved with combinatorial optimisation. Heuristic optimisation methods, such as genetic algorithms, greedy algorithms and further methods considered in the preparatory work, are used for this.